RMDP17
.........................................................................................
Introduction
The name RMDP17 refers to an exo-polysaccharide which is produced by Sphingomonas paucimobilis.
The structure if RMDP17 was determined by Jansson et al.  . It show a close similarity with the structure of rhamsan, as the only difference is on the uronic acid which is a 2-deoxy-glucuronic acid in RMDP17 instead of a glucuronic acid in Rhamsan. It has been shown that both optical rotation and differential scanning calorimetry measurements ascribe higher thermal stability to RMDP17 compared to rhamsan. . It show a close similarity with the structure of rhamsan, as the only difference is on the uronic acid which is a 2-deoxy-glucuronic acid in RMDP17 instead of a glucuronic acid in Rhamsan. It has been shown that both optical rotation and differential scanning calorimetry measurements ascribe higher thermal stability to RMDP17 compared to rhamsan.
a. 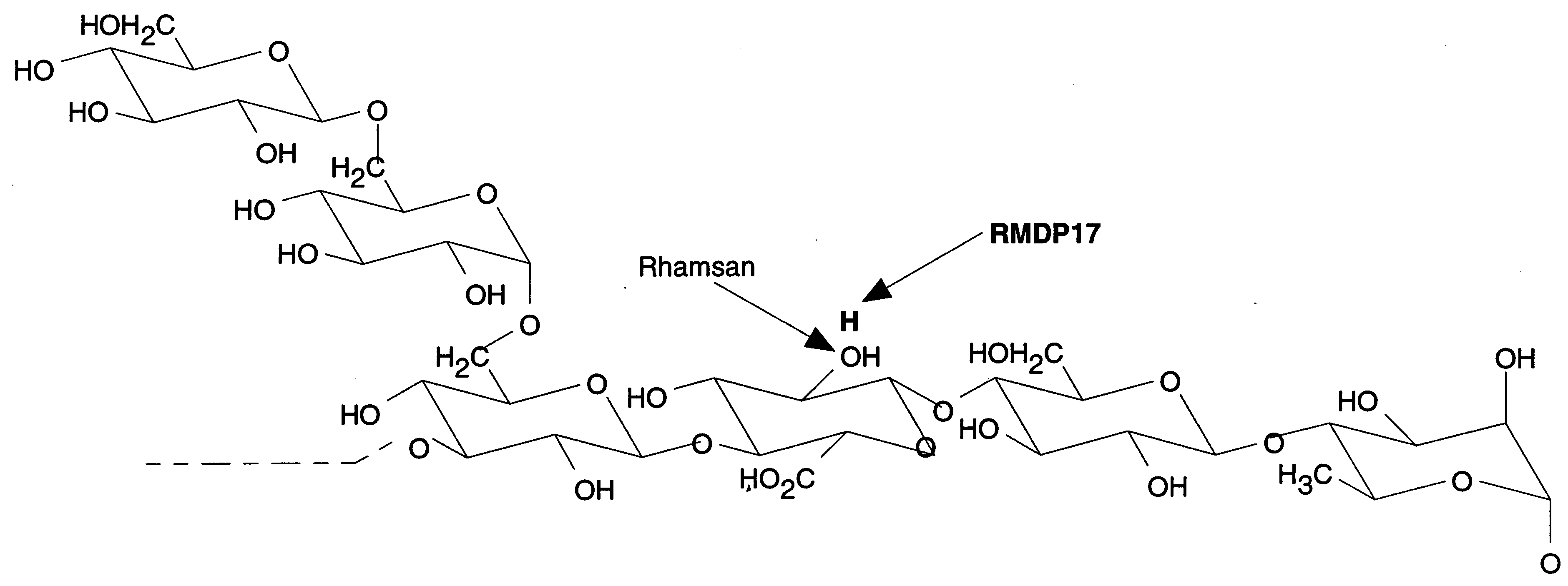
b. 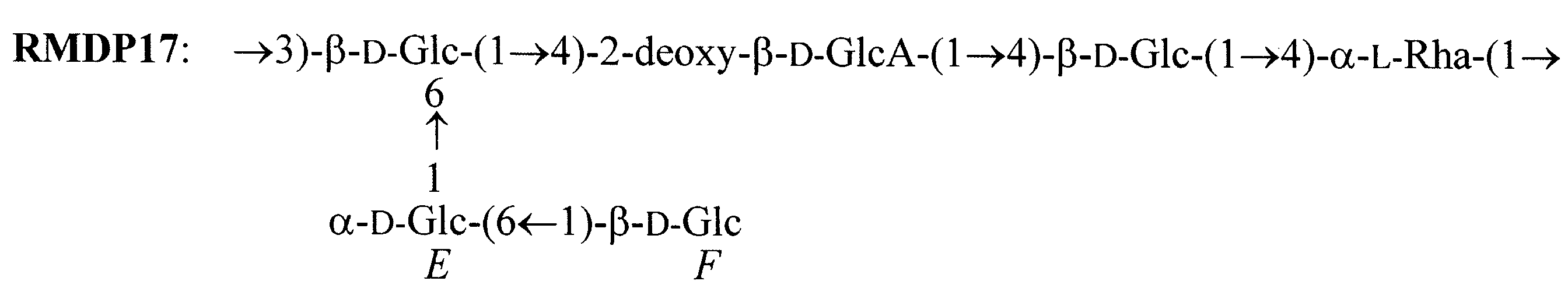
Fig. 1 Schematic representation of RMDP17 (a) Ring representation (b) Primary structure
X-ray diffraction pattern have been obtained from a well oriented fiber of the sodium salt of RMDP17 kept at 75% relative humidity. All the observed X-ray reflections can be indexed on a trigonal unit cell having dimensions of a = 17.6 Ang., c = 28.7 Å. From the measured fiber density of 1.53 it can be concluded that the unit cell accommodates six hexasaccharide repeats as well as a sodium ion and 11 water molecules. The meridional reflections on layer line 3 and 6 imply the occurrence of three-fold helix symmetry. The full analysis of the diffraction pattern, establishes the features of the structure as a half-staggered, threefold, left handed double helix of a pitch a 57.4 Å. The side chain of the branched polysaccharide is hydrogen bonded to the main-chain. Throughout their interactions with the carboxylate groups the sodium ions promote the associations of helices via water molecules. Within the unit cell, the two helices have an opposite polarity  . .
|








