A Database of Polysachharide 3D structures




| Home | |||||||
| About | |||||||
| User Guide | |||||||
| > GAG Structures | |||||||
| Methods | |||||||
| References | |||||||
| Wiki | |||||||
| Contact us | |||||||
|
** External Links **
|
||||||
|
|||||||
Discover Polysaccharides |
Glycosaminoglycans (GAGs)
......................................................................................... Glycosaminoglycan (GAGs) are linear polysaccharides present on all animal cell surfaces and in the extracellular matrix, where they are usually found to be attached covalently to core proteins to form the proteoglycan family. Each tissue produces specific repertoires of glycosaminoglycans (GAGs), some of which are known to bind and regulate chemokine activity. GAGs can be classified into four groups : the hyaluronic acid type, the chondroitin / dermatan sulfate type, the Heparan/Heparin type, and the Keratan type. They are acidic polysaccharides made of disaccharide repeating units (ranging from 40 to 100) which consists of uronic acid and hexosamines. Except for the hyaluronic acid, epimerization at the C-5 position of uronic acids, and N- and O- sulfation provide numerous sources of microheterogeneities. GAGs assume extended structures in aqueous solutions because of their strong hydrophilic nature based on their extensive sulfation, which is further exaggerated when they are covalently linked to core proteins. They hold a large number of water molecules in their molecular domain and occupy enormous hydrodynamic space in solution. A complementary remarkable property of the glycosaminoglycans, which is particularly significant in heparan sulfate and heparin, is their capability to specifically interact with a number of important growth factors and functional proteins. Such interactions are often crucial to the biological functions of these proteins.
Hyaluronic acid →4(–β-D-GlcA-1→3-β-D-GlcNAc-1→)n Chrondroitin / Dermatan →4(–β-D-GlcA-1→3-β-D-GalNAc-1→)n Heparan sulfate / Heparin →4(–β-D-GlcA-1→4-α-D-GlcN-R-1→)n Keratan sulfate →3(–β-D-Gal-1→3-β-D-GlcNAc-1→)n
Fig. 1 A proteoglycanglycan : a large molecular complex formed by the linking of many GAGs (composed of oligosaccharide chains attached to the core protein) through a long hyaluronate strand
Fig. 2 A schematic representation of a glycosaminoglycan (Mucopolysaccharide) its location and density
Fig. 3 A 2D representation of the Hyaluronan structure From X-ray investigation, seven polymorphs have been characterized which can be classified under three major categories that range form 4-fold to 3-fold left-handed helices. These are only some of the possible theoretical helical conformations that hyaluronan can adopt.
Fig. 4 A schematic drawing of hyaluronic acid segment. Each disaccharide repeat has the potential to participate in 4 hydrogen bonds (2 across the 1→4 linkage and 2 across the 1→3 linkage)
The 3-fold conformation is limited to a fully stretched value of 9.5 Å for the disaccharide repeat. The 4-fold conformation is more tolerant to stretching and upon external conditions such the nature of the cation, and/or the relative humidity, can adopt conformations such that the length of the disaccharide repeat may vary from full extension of 9.5 Å to 8.5 Å. Under the influence of potassium ions, a double helical structure may occur. Sodium Hyaluronate I anhydrous. The fiber diffraction diagram for sodium hyaluronate in its anhydrous form can be interpreted based on a 4-fold (43) helix symmetry
a. b. c. Fig. 5 A 3D representation of (a.) hyaluronate I sodium (b.) hyaluronate II sodium (c.) hyaluronate III sodium
The space group symmetry has been identified as P43212, and two helices passes through the unit cell. They are oriented in an anti-parallel fashion, into a tight packing anhydrous arrangement. Sodium being the only guest molecule interacts with the carboxylate group. It dispays an octahedral coordination. Anti-parallel and parallel helices interacts via hydrogen bonds, respectively O3H....O62 (2.59 Å) and O2H...O6 (2.37 Å) and O6H...O7 (2.79 Å). (Note the O2H...O6 distance of 2.37 Å is notably too short thereby indicated a minor flaw in the elaboration of the model).
Sodium Hyaluronate I hydrated. Upon control relative humidity at 75%, one observes an alteration of the unit cell and space group symmetry of the anhydrous form
Fig. 6 A 3D representation of the structure reported by Guss et al.
Potassium Hyaluronate I. Upon replacement of sodium per potassium, and while maintaing a relative humidity at 85% an X-ray fiber diffractogram that can be indexed in a tetragonal space, P43212 group occurs
Fig. 7 A 3D representation of the structure of Potassium Hyaluronate I
Potassium Hyaluronate II. Another fiber diffraction pattern can be found for potassium hyaluronate
These structural models have been confirmed throughout independant investigations
Sodium Hyaluronate III. This is another hydrated polymorph that is obtained when some of the fibers are prepared at 40°C with 90% relative humidity. Under these conditions, diffraction patterns are obtained, that correspond to the occurrence of 3-fold helix symmetry
Fig. 8 A 3D representation of the structure of Sodium Hyaluronate III
The unit cell of the P3121 space group encompasses two helices, one sodium ion per disaccharide and about 3.5 water molecules. The sodium ions have a total of 6 ligands and an octahedral coordination. The helices are aligned in an anti-parallel arrangement, and are associated via carboxylate...sodium...O2 (glucuronate) / O7 (carbonyl interactions), with a further stabilisation by an interchain hydrogen bond (O6-H... O3 (2.72 Å).
Calcium Hyaluronate. X-ray fiber diffractogram of calcium hyaluronate obtained at 75% relative humidity (and higher) can be interpreted on the basis of a 3-fold helix symmetry, occurring in a trigonal spce group, P3112, with a unit-cell having large dimensions. Fig. 9 A 3D representation of the structure of Calcium Hyaluronate
Potassium Hyaluronate III. When prepared at low pH (3.0 – 4.0) well oriented fibers of potassium hyaluronate correspond to a hemi or fully protonated state of the polysaccharide. The interpretation of the corresponding X-ray diffractogram
Fig. 10 A 3D representation of the structure of Potassium Hyaluronate III
The cation are regularly placed between the duplexes, and have been considered to be redundant for the stability and the survival of the double-helical structure. Inter double-helix hydrogen bonds are mediated between hydroxymethyl and N-acetly groups.
Rationalising the conformational versatility of hyaluronan chains Fig. 11 Helices of hyaluronic acid predicted by molecular modelling
Finally the feasibility of a double helix with anti-parallel strands for hyaluronan chains was evaluated and compared to the two models derived from X-ray experiments. The calculations demonstrated that the formation of a duplex by chain folding is possible; though marginally observed, the anti-parallel arrangement of the chains in the double helix is more favourable than the parallel one for hyaluronic acid. The energetic evaluation of the X-ray models, in comparison to the established theoretical model, indicated that the first model originally proposed by Sheehan and coworkers is not viable, and that the second one proposed by Arnott et al. is much more reasonable, with may be an over-evaluated hydrogen bond network.
The repeating unit of chondroitin is a disaccharide similar to that in hyaluronan, except that GalNAc replaces GlcNAc.
In the native state, the galactose residues are O-sulfated (either at position 4 or at position 6). Molecular modelling investigations
Sodium Chrondroitin 4-sulfate and Potassium Chondroitin 4-sulfate. For both salts X-ray analysis from fiber diffractogram has established the occurrence of a left-handed 3 fold helix with the sulfate groups pointing towards the periphery where they interact with one surrounding cation and water molecules
Fig. 13 A 3D representation of the structure of Sodium Chondroitin 4-sulphate
Fig. 14 A 3D representation of the structure of Potassium Chondroitin 4-sulphate
Sodium Chrondroitin 4-sulfate. The replacement of monovalent ions with divalent ions such as calcium induces a conformational transition of the chain from 3 fold helix to 2-fold helix symmetry
Dermatan 4 sulfate differs from chondroitin by having α-L-iduronate instead of β-D-glucuronate residues. As with the other members of the family of Glycosaminoglycan, dermatan sulfate displays polymorphism. Three distinct forms of molecular structure and packing arrangements have been identified from X-ray diffraction patterns. A first polymorph
Fig. 15 A 3D representation of the structure of Dermatan sulphate - allomorph I
A second polymorph displays a two-fold helix pitch of 18.8 Å and bears some similraity to the structure of calcium chloride chondroitin 4-sulfate.
Fig. 16 A 3D representation of the structure of Dermatan sulphate - allomorph II A fairly novel form was found for a third polymorph which was identified as a right-handed helix with 83 symmetry (disaccharide extension of 9.2 Å) made up of a tetrasaccharide in which two slightly different disaccharides repeat. It this model, chains are packed in an anti-parallel fashion in the P43212 tetragonal space group. Fig. 17 A 3D representation of the structure of Dermatan sulphate - allomorph III X-ray diffraction patterns from stretched films of keratan 6-sulfate show only continuous intensities from which one can identify meridional reflections on even layers indicating the occurrence of a two-fold helix with an extension of 9.45 Å per disaccharide
Fig. 18 A 3D representation of the structure of Keratan 6-sulphate
Heparin is the most therapeutically (and commercially) important member of the GAG family. For the first 30 years of its clinical use, not much was known about its structure except that iy was composed of glucosamine and uronic acid, with heavy sulfate substitution. Heparin has much higher specific anticoagulant activity than any other sulfated polysaccharides. The main structural components of heparin were established as containing N- and 6-O-sulfated α-D-glucosamine, 2-O-sulfated a-l iduronic acid along with N-acetyl D-glucosamine and β-D-glucuronic acid
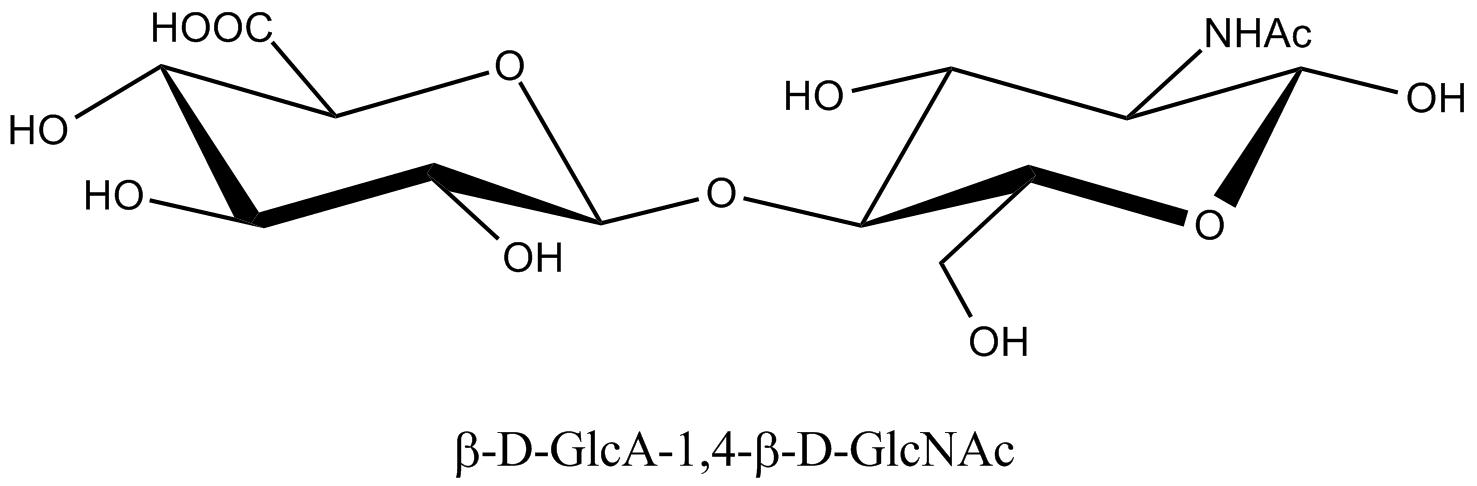
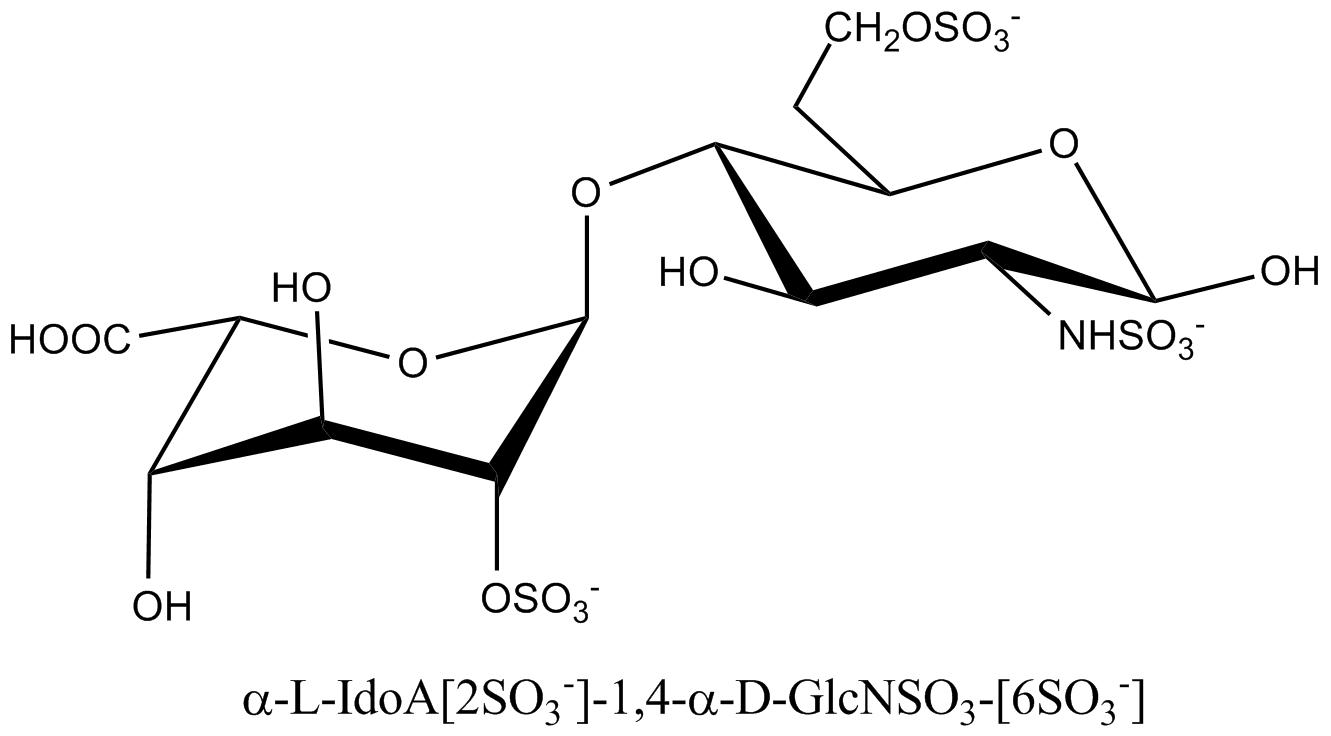
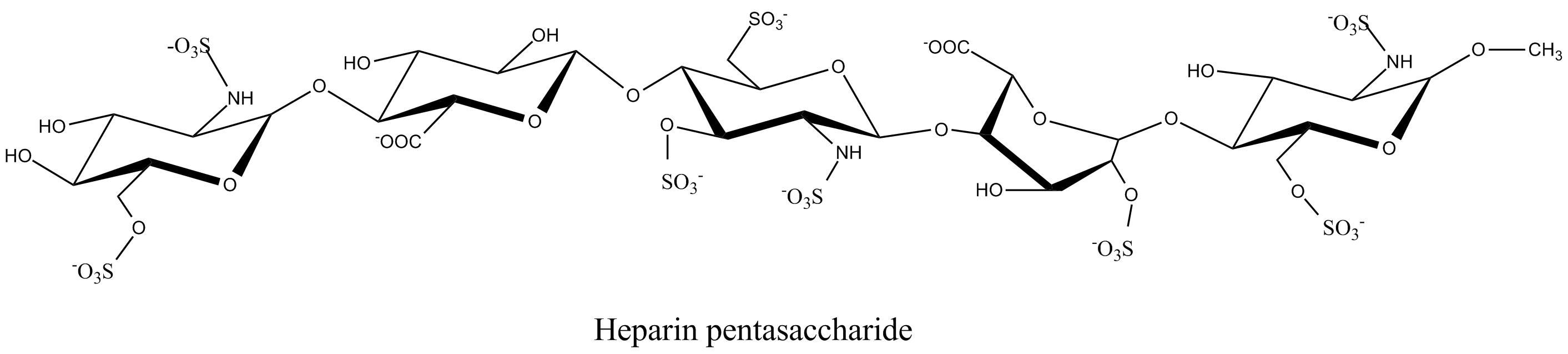
a. b. c. Fig. 19 Chemical structure representations of heparin and heparin sulphate (a) Precursor polysaccharide for both heparin & heparan sulphate, β-D-GlcA-1,4-β-D-GlcNAc, which is also a predominant structure in heparan sulphate. (b) Post-polymerization, enzymes remove (i) remove N-acetyl and replace it with N-sulphate (ii) epimerize β-D-glucuronate residues at the resucing side of N-sulphate to α-L-iduronate (iii) introduce 2-O-sulphate and (iv) introduce 6-O-sulphate. The final structure from all these changes results of these transformations leads to the formation of the main repeating unit of heparin α-L-IdoA[2SO3-]-(1,4)-α-D-GlcNSO3-[6SO3-] (c) The pentasaccharide in heparin α-D-GlcNAc[6SO3-]-(1→4)-α-D-GlcA-(1→4)-α-D-GlcNSO3-[3, 6-diSO3-]-(1→4)-α-L-IdoA[2SO3-]-(1→4)-α-D-GlcNSO3-[6SO3-] which is the minimal structure with high affinity for anti-thrombin, the serine protease in plasma which inhibits several coagulation enzymes.
Fig. 20 X-ray fibre-diffraction patterns obtained from heparan sulphate The β-D-glucuronic and &alpha-D-N-acetyl glucosamine makes up most of the structure of heparan sulfate, interrupted by N-sulfated iduronate – containing sequences, which have been denotated as S-regions. It is through these heparin-like S-domains that heparan sulfate interacts with proteins. Conformational studies of heparin and molecular modeling have shown that the polysaccharide chain has a relatively well-defined shape, regardless of its patter, of sulfate substitutions. Nevertheless, sequence determines the three-dimensional structure of the chains, at more than one level. Detailed sequences of highly sulfated regions may influence affinity for specific interactions with proteins. In addition, the length and spacing of these highly sulfated sequences which are separated by unsulfated domains may also be crucial components. The sulfated S-domains offer space for internal dynamics of the conformationally flexible iduronate rings. New studies of the relative degrees of flexibility of sulfated and unsulfated domains lead to an overall model of heparin/heparan sulfate in which proteins-dinding, highly sulfated S-domains with well-defined conformations are separated by more flexible domains
|